Coffea Processors
This is a rendered copy of processor.ipynb. You can optionally run it interactively on binder at this link
Coffea relies mainly on uproot to provide access to ROOT files for analysis. As a usual analysis will involve processing tens to thousands of files, totalling gigabytes to terabytes of data, there is a certain amount of work to be done to build a parallelized framework to process the data in a reasonable amount of time. Of course, one can work directly within uproot to achieve this, as we’ll show in the beginning, but coffea provides the
coffea.processor module, which allows users to worry just about the actual analysis code and not about how to implement efficient parallelization, assuming that the parallization is a trivial map-reduce operation (e.g. filling histograms and adding them together). The module provides the following key features:
A
ProcessorABCabstract base class that can be derived from to implement the analysis code;A NanoEvents interface to the arrays being read from the TTree as inputs;
A generic
accumulate()utility to reduce the outputs to a single result, as showin in the accumulators notebook tutorial; andA set of parallel executors to access multicore processing or distributed computing systems such as Dask, Parsl, Spark, WorkQueue, and others.
Let’s start by writing a simple processor class that reads some CMS open data and plots a dimuon mass spectrum. We’ll start by copying the ProcessorABC skeleton and filling in some details:
Remove
flag, as we won’t use itAdding a new histogram for \(m_{\mu \mu}\)
Building a Candidate record for muons, since we will read it with
BaseSchemainterpretation (the files used here could be read withNanoAODSchemabut we want to show how to build vector objects from other TTree formats)Calculating the dimuon invariant mass
[1]:
import awkward as ak
from coffea import processor
from coffea.nanoevents.methods import candidate
import hist
class MyProcessor(processor.ProcessorABC):
def __init__(self):
pass
def process(self, events):
dataset = events.metadata['dataset']
muons = ak.zip(
{
"pt": events.Muon_pt,
"eta": events.Muon_eta,
"phi": events.Muon_phi,
"mass": events.Muon_mass,
"charge": events.Muon_charge,
},
with_name="PtEtaPhiMCandidate",
behavior=candidate.behavior,
)
h_mass = (
hist.Hist.new
.StrCat(["opposite", "same"], name="sign")
.Log(1000, 0.2, 200., name="mass", label="$m_{\mu\mu}$ [GeV]")
.Int64()
)
cut = (ak.num(muons) == 2) & (ak.sum(muons.charge, axis=1) == 0)
# add first and second muon in every event together
dimuon = muons[cut][:, 0] + muons[cut][:, 1]
h_mass.fill(sign="opposite", mass=dimuon.mass)
cut = (ak.num(muons) == 2) & (ak.sum(muons.charge, axis=1) != 0)
dimuon = muons[cut][:, 0] + muons[cut][:, 1]
h_mass.fill(sign="same", mass=dimuon.mass)
return {
dataset: {
"entries": len(events),
"mass": h_mass,
}
}
def postprocess(self, accumulator):
pass
If we were to just use bare uproot to execute this processor, we could do that with the following example, which:
Opens a CMS open data file
Creates a NanoEvents object using
BaseSchema(roughly equivalent to the output ofuproot.lazy)Creates a
MyProcessorinstanceRuns the
process()function, which returns our accumulators
[2]:
import uproot
from coffea.nanoevents import NanoEventsFactory, BaseSchema
filename = "root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod/Run2012B_DoubleMuParked.root"
file = uproot.open(filename)
events = NanoEventsFactory.from_root(
file,
entry_stop=10000,
metadata={"dataset": "DoubleMuon"},
schemaclass=BaseSchema,
).events()
p = MyProcessor()
out = p.process(events)
out
[2]:
{'DoubleMuon': {'entries': 10000,
'mass': Hist(
StrCategory(['opposite', 'same'], name='sign', label='sign'),
Regular(1000, 0.2, 200, transform=log, name='mass', label='$m_{\\mu\\mu}$ [GeV]'),
storage=Int64()) # Sum: 4939.0 (4951.0 with flow)}}
[3]:
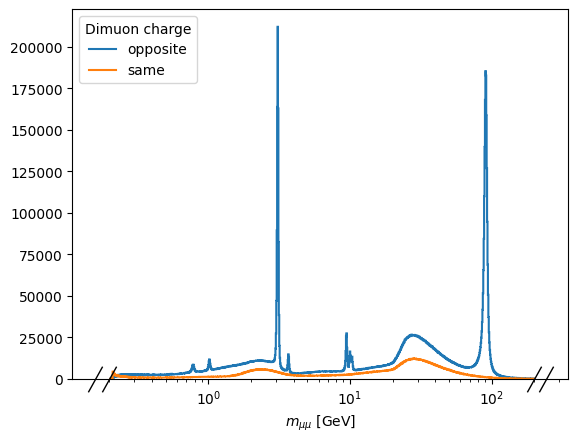
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
out["DoubleMuon"]["mass"].plot1d(ax=ax)
ax.set_xscale("log")
ax.legend(title="Dimuon charge")
[3]:
<matplotlib.legend.Legend at 0x7f083c0c6310>
One could expand on this code to run over several chunks of the file, setting entry_start and entry_stop as appropriate. Then, several datasets could be processed by iterating over several files. However, the processor Runner can help with this! One lists the datasets and corresponding files, the processor they want to run, and which executor they want to use. Available executors derive from ExecutorBase and
are listed here. Since these files are very large, we limit to just reading the first few chunks of events from each dataset with maxchunks.
[4]:
fileset = {
'DoubleMuon': [
'root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod/Run2012B_DoubleMuParked.root',
'root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod/Run2012C_DoubleMuParked.root',
],
'ZZ to 4mu': [
'root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod/ZZTo4mu.root'
]
}
iterative_run = processor.Runner(
executor = processor.IterativeExecutor(compression=None),
schema=BaseSchema,
maxchunks=10,
)
out = iterative_run(
fileset,
treename="Events",
processor_instance=MyProcessor(),
)
out
/opt/conda/lib/python3.8/site-packages/awkward/_connect/_numpy.py:195: RuntimeWarning: invalid value encountered in sqrt result = getattr(ufunc, method)(
[4]:
{'ZZ to 4mu': {'entries': 999380,
'mass': Hist(
StrCategory(['opposite', 'same'], name='sign', label='sign'),
Regular(1000, 0.2, 200, transform=log, name='mass', label='$m_{\\mu\\mu}$ [GeV]'),
storage=Int64()) # Sum: 192074.0 (192500.0 with flow)},
'DoubleMuon': {'entries': 1000560,
'mass': Hist(
StrCategory(['opposite', 'same'], name='sign', label='sign'),
Regular(1000, 0.2, 200, transform=log, name='mass', label='$m_{\\mu\\mu}$ [GeV]'),
storage=Int64()) # Sum: 519403.0 (520002.0 with flow)}}
Now, if we want to use more than a single core on our machine, we simply change IterativeExecutor for FuturesExecutor, which uses the python concurrent.futures standard library. We can then set the most interesting argument to the FuturesExecutor: the number of
cores to use (2):
[5]:
futures_run = processor.Runner(
executor = processor.FuturesExecutor(compression=None, workers=2),
schema=BaseSchema,
maxchunks=10,
)
out = futures_run(
fileset,
"Events",
processor_instance=MyProcessor()
)
out
[5]:
{'ZZ to 4mu': {'entries': 999380,
'mass': Hist(
StrCategory(['opposite', 'same'], name='sign', label='sign'),
Regular(1000, 0.2, 200, transform=log, name='mass', label='$m_{\\mu\\mu}$ [GeV]'),
storage=Int64()) # Sum: 192074.0 (192500.0 with flow)},
'DoubleMuon': {'entries': 1000560,
'mass': Hist(
StrCategory(['opposite', 'same'], name='sign', label='sign'),
Regular(1000, 0.2, 200, transform=log, name='mass', label='$m_{\\mu\\mu}$ [GeV]'),
storage=Int64()) # Sum: 519403.0 (520002.0 with flow)}}
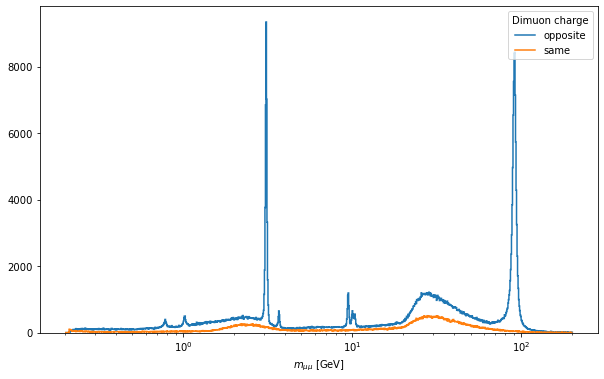
Hopefully this ran faster than the previous cell, but that may depend on how many cores are available on the machine you are running this notebook and your connection to eospublic.cern.ch. At least the output will be prettier now:
[6]:
fig, ax = plt.subplots(figsize=(10, 6))
out["DoubleMuon"]["mass"].plot1d(ax=ax)
ax.set_xscale("log")
ax.legend(title="Dimuon charge")
[6]:
<matplotlib.legend.Legend at 0x7f08123c89d0>
Getting fancy
Let’s flesh out this analysis into a 4-muon analysis, searching for diboson events:
[8]:
from collections import defaultdict
import numba
@numba.njit
def find_4lep(events_leptons, builder):
"""Search for valid 4-lepton combinations from an array of events * leptons {charge, ...}
A valid candidate has two pairs of leptons that each have balanced charge
Outputs an array of events * candidates {indices 0..3} corresponding to all valid
permutations of all valid combinations of unique leptons in each event
(omitting permutations of the pairs)
"""
for leptons in events_leptons:
builder.begin_list()
nlep = len(leptons)
for i0 in range(nlep):
for i1 in range(i0 + 1, nlep):
if leptons[i0].charge + leptons[i1].charge != 0:
continue
for i2 in range(nlep):
for i3 in range(i2 + 1, nlep):
if len({i0, i1, i2, i3}) < 4:
continue
if leptons[i2].charge + leptons[i3].charge != 0:
continue
builder.begin_tuple(4)
builder.index(0).integer(i0)
builder.index(1).integer(i1)
builder.index(2).integer(i2)
builder.index(3).integer(i3)
builder.end_tuple()
builder.end_list()
return builder
class FancyDimuonProcessor(processor.ProcessorABC):
def process(self, events):
dataset_axis = hist.axis.StrCategory([], growth=True, name="dataset", label="Primary dataset")
mass_axis = hist.axis.Regular(300, 0, 300, name="mass", label=r"$m_{\mu\mu}$ [GeV]")
pt_axis = hist.axis.Regular(300, 0, 300, name="pt", label=r"$p_{T,\mu}$ [GeV]")
h_nMuons = hist.Hist(
dataset_axis,
hist.axis.IntCategory(range(6), name="nMuons", label="Number of good muons"),
storage="weight", label="Counts",
)
h_m4mu = hist.Hist(dataset_axis, mass_axis, storage="weight", label="Counts")
h_mZ1 = hist.Hist(dataset_axis, mass_axis, storage="weight", label="Counts")
h_mZ2 = hist.Hist(dataset_axis, mass_axis, storage="weight", label="Counts")
h_ptZ1mu1 = hist.Hist(dataset_axis, pt_axis, storage="weight", label="Counts")
h_ptZ1mu2 = hist.Hist(dataset_axis, pt_axis, storage="weight", label="Counts")
cutflow = defaultdict(int)
dataset = events.metadata['dataset']
muons = ak.zip({
"pt": events.Muon_pt,
"eta": events.Muon_eta,
"phi": events.Muon_phi,
"mass": events.Muon_mass,
"charge": events.Muon_charge,
"softId": events.Muon_softId,
"isolation": events.Muon_pfRelIso03_all,
}, with_name="PtEtaPhiMCandidate", behavior=candidate.behavior)
# make sure they are sorted by transverse momentum
muons = muons[ak.argsort(muons.pt, axis=1)]
cutflow['all events'] += len(muons)
# impose some quality and minimum pt cuts on the muons
muons = muons[
muons.softId
& (muons.pt > 5)
& (muons.isolation < 0.2)
]
cutflow['at least 4 good muons'] += ak.sum(ak.num(muons) >= 4)
h_nMuons.fill(dataset=dataset, nMuons=ak.num(muons))
# reduce first axis: skip events without enough muons
muons = muons[ak.num(muons) >= 4]
# find all candidates with helper function
fourmuon = find_4lep(muons, ak.ArrayBuilder()).snapshot()
if ak.all(ak.num(fourmuon) == 0):
# skip processing as it is an EmptyArray
return {
'nMuons': h_nMuons,
'cutflow': {dataset: cutflow},
}
fourmuon = [muons[fourmuon[idx]] for idx in "0123"]
fourmuon = ak.zip({
"z1": ak.zip({
"lep1": fourmuon[0],
"lep2": fourmuon[1],
"p4": fourmuon[0] + fourmuon[1],
}),
"z2": ak.zip({
"lep1": fourmuon[2],
"lep2": fourmuon[3],
"p4": fourmuon[2] + fourmuon[3],
}),
})
cutflow['at least one candidate'] += ak.sum(ak.num(fourmuon) > 0)
# require minimum dimuon mass
fourmuon = fourmuon[(fourmuon.z1.p4.mass > 60.) & (fourmuon.z2.p4.mass > 20.)]
cutflow['minimum dimuon mass'] += ak.sum(ak.num(fourmuon) > 0)
# choose permutation with z1 mass closest to nominal Z boson mass
bestz1 = ak.singletons(ak.argmin(abs(fourmuon.z1.p4.mass - 91.1876), axis=1))
fourmuon = ak.flatten(fourmuon[bestz1])
h_m4mu.fill(
dataset=dataset,
mass=(fourmuon.z1.p4 + fourmuon.z2.p4).mass,
)
h_mZ1.fill(
dataset=dataset,
mass=fourmuon.z1.p4.mass,
)
h_mZ2.fill(
dataset=dataset,
mass=fourmuon.z2.p4.mass,
)
h_ptZ1mu1.fill(
dataset=dataset,
pt=fourmuon.z1.lep1.pt,
)
h_ptZ1mu2.fill(
dataset=dataset,
pt=fourmuon.z1.lep2.pt,
)
return {
'nMuons': h_nMuons,
'mass': h_m4mu,
'mass_z1': h_mZ1,
'mass_z2': h_mZ2,
'pt_z1_mu1': h_ptZ1mu1,
'pt_z1_mu2': h_ptZ1mu2,
'cutflow': {dataset: cutflow},
}
def postprocess(self, accumulator):
pass
[9]:
import time
tstart = time.time()
fileset = {
'DoubleMuon': [
'root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod/Run2012B_DoubleMuParked.root',
'root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod/Run2012C_DoubleMuParked.root',
],
'ZZ to 4mu': [
'root://eospublic.cern.ch//eos/root-eos/cms_opendata_2012_nanoaod/ZZTo4mu.root'
]
}
run = processor.Runner(
executor = processor.FuturesExecutor(compression=None, workers=4),
schema=BaseSchema,
chunksize=100_000,
# maxchunks=10, # total 676 chunks
)
output = run(
fileset,
"Events",
processor_instance=FancyDimuonProcessor(),
)
elapsed = time.time() - tstart
print(output)
{'nMuons': Hist(
StrCategory(['ZZ to 4mu', 'DoubleMuon'], growth=True, name='dataset', label='Primary dataset'),
IntCategory([0, 1, 2, 3, 4, 5], name='nMuons', label='Number of good muons'),
storage=Weight()) # Sum: WeightedSum(value=6.76277e+07, variance=6.76277e+07) (WeightedSum(value=6.76279e+07, variance=6.76279e+07) with flow), 'mass': Hist(
StrCategory(['ZZ to 4mu', 'DoubleMuon'], growth=True, name='dataset', label='Primary dataset'),
Regular(300, 0, 300, name='mass', label='$m_{\\mu\\mu}$ [GeV]'),
storage=Weight()) # Sum: WeightedSum(value=65472, variance=65472) (WeightedSum(value=82352, variance=82352) with flow), 'mass_z1': Hist(
StrCategory(['ZZ to 4mu', 'DoubleMuon'], growth=True, name='dataset', label='Primary dataset'),
Regular(300, 0, 300, name='mass', label='$m_{\\mu\\mu}$ [GeV]'),
storage=Weight()) # Sum: WeightedSum(value=82303, variance=82303) (WeightedSum(value=82352, variance=82352) with flow), 'mass_z2': Hist(
StrCategory(['ZZ to 4mu', 'DoubleMuon'], growth=True, name='dataset', label='Primary dataset'),
Regular(300, 0, 300, name='mass', label='$m_{\\mu\\mu}$ [GeV]'),
storage=Weight()) # Sum: WeightedSum(value=81873, variance=81873) (WeightedSum(value=82352, variance=82352) with flow), 'pt_z1_mu1': Hist(
StrCategory(['ZZ to 4mu', 'DoubleMuon'], growth=True, name='dataset', label='Primary dataset'),
Regular(300, 0, 300, name='pt', label='$p_{T,\\mu}$ [GeV]'),
storage=Weight()) # Sum: WeightedSum(value=82351, variance=82351) (WeightedSum(value=82352, variance=82352) with flow), 'pt_z1_mu2': Hist(
StrCategory(['ZZ to 4mu', 'DoubleMuon'], growth=True, name='dataset', label='Primary dataset'),
Regular(300, 0, 300, name='pt', label='$p_{T,\\mu}$ [GeV]'),
storage=Weight()) # Sum: WeightedSum(value=82170, variance=82170) (WeightedSum(value=82352, variance=82352) with flow), 'cutflow': {'ZZ to 4mu': defaultdict(<class 'int'>, {'all events': 1499064, 'at least 4 good muons': 143618, 'at least one candidate': 143055, 'minimum dimuon mass': 81867}), 'DoubleMuon': defaultdict(<class 'int'>, {'all events': 66128870, 'at least 4 good muons': 8289, 'at least one candidate': 3849, 'minimum dimuon mass': 485})}}
[10]:
nevt = output['cutflow']['ZZ to 4mu']['all events'] + output['cutflow']['DoubleMuon']['all events']
print("Events/s:", nevt / elapsed)
Events/s: 153664.49672142894
What follows is just us looking at the output, you can execute it if you wish
[11]:
# scale ZZ simulation to expected yield
lumi = 11.6 # 1/fb
zzxs = 7200 * 0.0336**2 # approximate 8 TeV ZZ(4mu)
nzz = output['cutflow']['ZZ to 4mu']['all events']
scaled = {}
for name, h in output.items():
if isinstance(h, hist.Hist):
scaled[name] = h.copy()
scaled[name].view()[0, :] *= lumi * zzxs / nzz
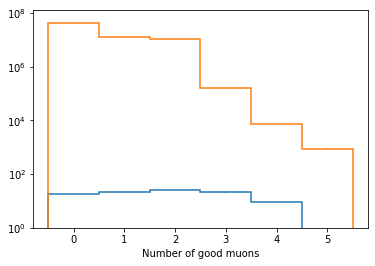
[12]:
fig, ax = plt.subplots()
scaled['nMuons'].plot1d(ax=ax, overlay='dataset')
ax.set_yscale('log')
ax.set_ylim(1, None)
[12]:
(1, 122819923.5463199)
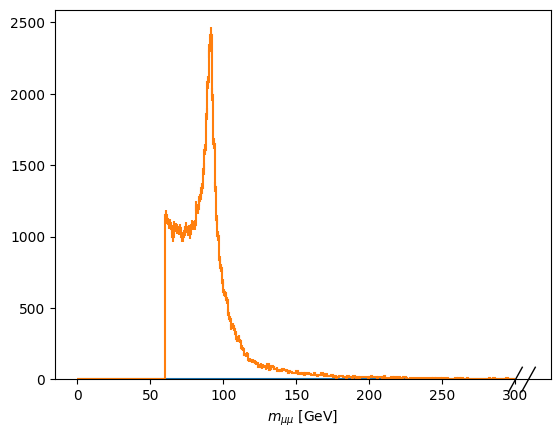
[13]:
fig, ax = plt.subplots()
scaled['mass'][:, ::hist.rebin(4)].plot1d(ax=ax, overlay='dataset');
[14]:
fig, ax = plt.subplots()
scaled['mass_z1'].plot1d(ax=ax, overlay='dataset');
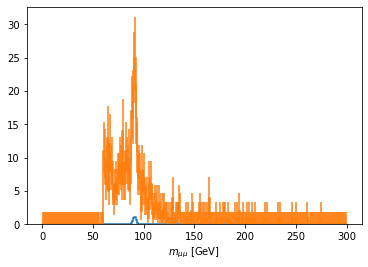
[15]:
fig, ax = plt.subplots()
scaled['mass_z2'].plot1d(ax=ax, overlay='dataset')
ax.set_xlim(2, 300)
# ax.set_xscale('log')
[15]:
(2.0, 300.0)
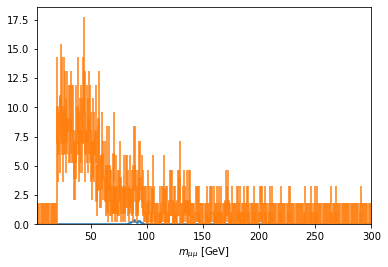
[16]:
fig, ax = plt.subplots()
scaled['pt_z1_mu1'].plot1d(ax=ax, overlay='dataset');
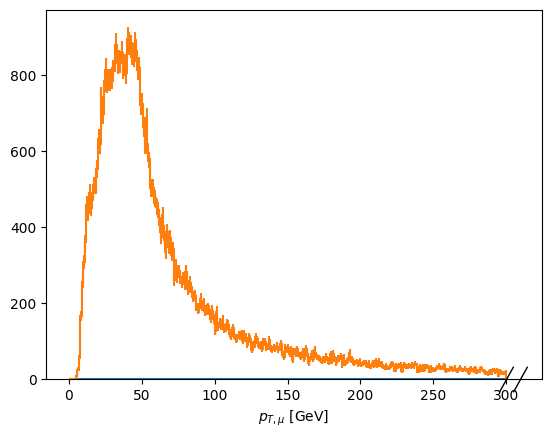
[17]:
fig, ax = plt.subplots()
scaled['pt_z1_mu2'].plot1d(ax=ax, overlay='dataset');
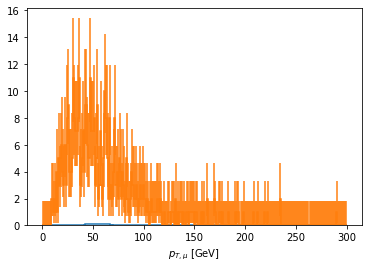
[ ]: